miRNA是非编码调控小分子,通过抑制基因表达发挥作用,前体pri-miRNA、pre-miRNA通过核内的drosa酶作用,由核孔复合体进入至胞浆中,在通过Dicer酶的剪切,形成成熟的miRNA,AGO2可介导miRNA结合至mRNA的3’UTR,进而对基因的表达进行抑制,可在mRNA、蛋白水平发生降解作用
我们对数据的整理,通过数据库查询分类、数据库功能及用途、示例结合分析、数据库优化等这四大项,进行阐述和演示数据库的查询和使用,希望对您的实验项目有所帮助
数据库查询分类
① microRNA.org:http://www.microrna.org/microrna/home.do
② TargetScan:http://www.targetscan.org/vert_70/
③ starbase: http://starbase.sysu.edu.cn/
④ Tarbase: http://www.tarbase.com/
⑤ deepBase: http://deepbase.sysu.edu.cn/
⑥ miRanda: http://www.microrna.org/microrna/home.do
⑦ RNAhybrid:https://bibiserv.cebitec.uni-bielefeld.de/
⑧ CoGeMiR:http://cogemir.tigem.it/
数据库功能及用途(常用数据库)
1. microRNA.org:
Predicted microRNA targets & target downregulation scores. Experimentally observed expression patterns
数据库功能:
1.
mirSVR predicted target site scoring method: Comprehensive modeling of
microRNA targets predicts functional non-conserved and non-canonical
sites
2. microRNA target predictions: The microRNA.org resource: targets and expression.
3. miRanda application: Human MicroRNA targets.
4. miRanda algorithm: MicroRNA targets in Drosophila.
2. TargetScan:
Search for predicted microRNA targets in mammals

数据库功能:
Search for predicted microRNA targets in mammals
3. starbase:
CLIP-Seq, Pan-Cancer. Portal, Visualize, Analyze, Discover.
数据库功能:
1.
starBase is designed for decoding Pan-Cancer and Interaction Networks
of lncRNAs, miRNAs, competing endogenous RNAs(ceRNAs), RNA-binding
proteins (RBPs) and mRNAs from large-scale CLIP-Seq (HITS-CLIP,
PAR-CLIP, iCLIP, CLASH) data and tumor samples (14 cancer types,
>6000 samples).
2. starBase
is also developed for deciphering Protein-RNA and miRNA-target
interactions, such as protein-lncRNA, protein-sncRNA, protein-mRNA,
protein-pseudogene, miRNA-lncRNA, miRNA-mRNA, miRNA-circRNA,
miRNA-pseudogene, miRNA-sncRNA interactions and ceRNA networks from 108
CLIP-Seq (HITS-CLIP, PAR-CLIP, iCLIP, CLASH) datasets.
3.
starBase provides miRFunction and ceRNAFunction web tools to predict
the function of ncRNAs (miRNAs, lncRNAs, pseduogenes) and protein-coding
genes from the miRNA-mediated (ceRNA) regulatory networks
示例结合分析:
microRNA.org:
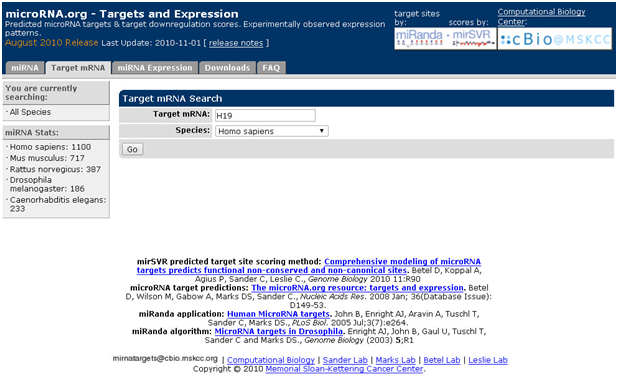
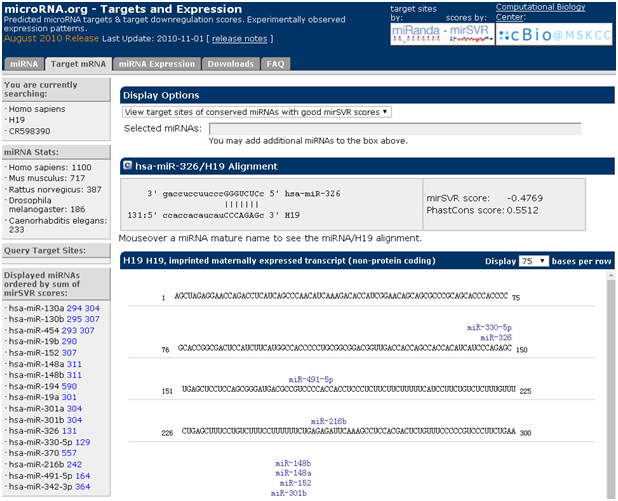
预测已知mRNA结合的miRNA:以H19为例
1. 打开主页面
2. 点击“target mRNA”,对话框输入h19,选择种属
3. 点击“GO”,即可获取得到预测结合的miRNA
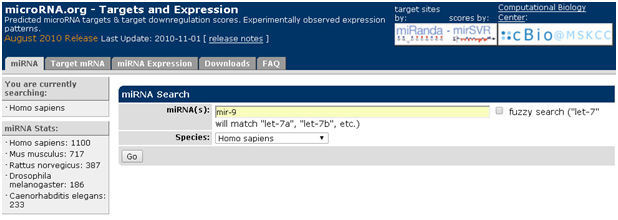
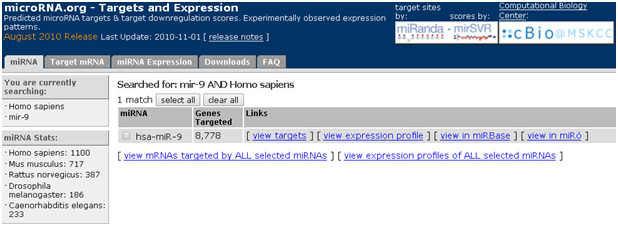
已知miRNA,查询该miRNA调控的mRNA:以mir-9为例
1. 打开主页面
2. 点击“miRNA”,对话框输入miR-9,选择种属
3. 点击“go”,即可获取miR-9调控的mRN
数据库优化:
microRNA.org是一款预测可以预测miRNA的靶标mRNA,以及引起靶标基因下调的得分值,以及miRNA的表达情况,也可以预测mRNA结合的miRNA,无法预测得到miRNA的调控网络
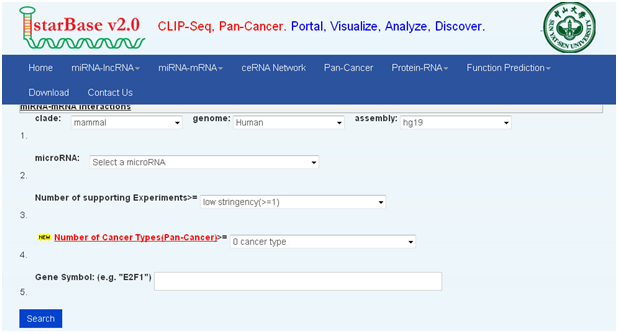
2. starBase:以mir-9为例
①打开主页面
②点击“miRNA-mRNA”,点击(两种查询:miRNA-mRNA intersections、miRNA-mRNA interactions)
③这里我们演示miRNA-mRNA interactions的示例
![]()
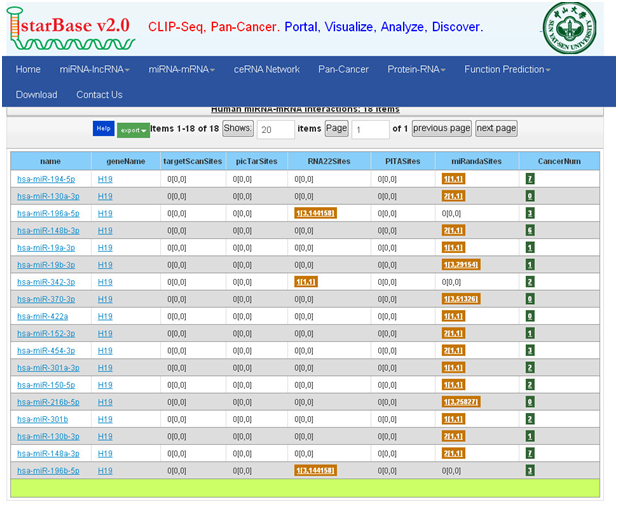
数据库优化:
Starbase可查询miRNA、LncRNA、mRNA相互调控和结合的预测,存储了大量的互作网络结合的miRNA,ceRNA的竞争结合,RNA蛋白结合等数据