# 清除当前环境中的变量
rm(list=ls())
# 设置工作路径
setwd("C:/Users/Dell/Desktop/")
# 加载所需的R包
library(ggplot2 )
library(pheatmap )
library(reshape2 )
# 读取测试数据
data <- read.table("test.txt",header = T, row.names = 1,check.names = F)
# 查看数据基本信息
head(data)
## Stage1_R1 Stage1_R2 Stage2_R1 Stage2_R2 Stage3_R1
## Unigene0001 -1.1777172 -1.036102 0.8423829 1.3458754 0.1080678
## Unigene0002 1.0596877 1.490939 -0.7663244 -0.6255567 -0.5333080
## Unigene0003 0.9206594 1.575844 -0.7861697 -0.3860003 -0.5501094
## Unigene0004 -1.3553173 -1.145970 0.2097526 0.7059886 0.9516353
## Unigene0005 1.0134516 1.445897 -0.9705129 -0.8560422 -0.2556562
## Unigene0006 0.8675939 1.575735 -1.0120718 -0.5856459 -0.2821991
## Stage3_R2
## Unigene0001 -0.08250721
## Unigene0002 -0.62543728
## Unigene0003 -0.77422398
## Unigene0004 0.63391053
## Unigene0005 -0.37713783
## Unigene0006 -0.56341216
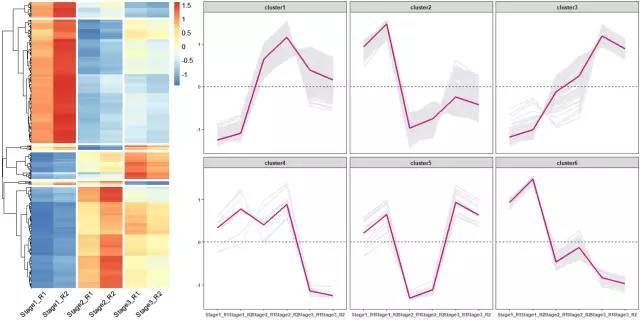
# 使用pheatmap绘制基因表达热图,并进行层次聚类分成不同的cluster
p <- pheatmap(data, show_rownames = F, cellwidth =40, cluster_cols = F,
cutree_rows = 6,gaps_col = c(2,4,6), angle_col = 45,fontsize = 12)
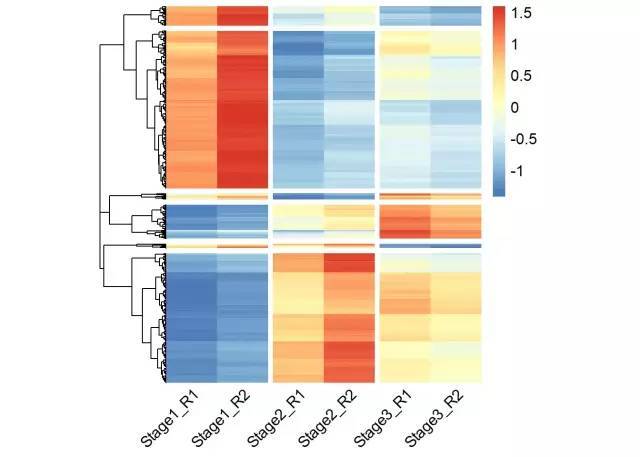
# 获取聚类后的基因顺序
row_cluster = cutree(p$tree_row,k=6)
# 对聚类后的数据进行重新排序
newOrder = data[p$tree_row$order,]
newOrder[,ncol(newOrder)+1]= row_cluster[match(rownames(newOrder),names(row_cluster))]
colnames(newOrder)[ncol(newOrder)]="Cluster"
# 查看重新排序后的数据
head(newOrder)
## Stage1_R1 Stage1_R2 Stage2_R1 Stage2_R2 Stage3_R1 Stage3_R2
## Unigene0604 0.8097531 1.403759 -0.2668053 0.17819117 -0.9811268 -1.143771
## Unigene0262 0.8453759 1.408372 -0.2802646 0.12312391 -0.9767547 -1.119853
## Unigene0069 0.8279061 1.428306 -0.3124647 0.12820543 -0.9524584 -1.119494
## Unigene0219 0.8536163 1.423168 -0.3082219 0.09583306 -0.9584284 -1.105967
## Unigene0116 0.8282198 1.491489 -0.4344344 0.05187827 -0.8641523 -1.073000
## Unigene0297 0.8008572 1.459959 -0.3661415 0.13242699 -0.9111229 -1.115978
## Cluster
## Unigene0604 6
## Unigene0262 6
## Unigene0069 6
## Unigene0219 6
## Unigene0116 6
## Unigene0297 6
# 查看聚类后cluster的基本信息
unique(newOrder$Cluster)
## [1] 6 2 5 3 4 1
table(newOrder$Cluster)
##
## 1 2 3 4 5 6
## 258 314 68 9 12 39
# 将新排序后的数据保存输出
newOrder$Cluster = paste0("cluster",newOrder$Cluster)
write.table(newOrder, "expr_DE.pheatmap.cluster.txt",sep="t",quote = F,row.names = T,col.names = T)
# 绘制每个cluster的基因聚类趋势图
newOrder$gene = rownames(newOrder)
head(newOrder)
## Stage1_R1 Stage1_R2 Stage2_R1 Stage2_R2 Stage3_R1 Stage3_R2
## Unigene0604 0.8097531 1.403759 -0.2668053 0.17819117 -0.9811268 -1.143771
## Unigene0262 0.8453759 1.408372 -0.2802646 0.12312391 -0.9767547 -1.119853
## Unigene0069 0.8279061 1.428306 -0.3124647 0.12820543 -0.9524584 -1.119494
## Unigene0219 0.8536163 1.423168 -0.3082219 0.09583306 -0.9584284 -1.105967
## Unigene0116 0.8282198 1.491489 -0.4344344 0.05187827 -0.8641523 -1.073000
## Unigene0297 0.8008572 1.459959 -0.3661415 0.13242699 -0.9111229 -1.115978
## Cluster gene
## Unigene0604 cluster6 Unigene0604
## Unigene0262 cluster6 Unigene0262
## Unigene0069 cluster6 Unigene0069
## Unigene0219 cluster6 Unigene0219
## Unigene0116 cluster6 Unigene0116
## Unigene0297 cluster6 Unigene0297
library(reshape2)
# 将短数据格式转换为长数据格式
data_new = melt(newOrder)
## Using Cluster, gene as id variables
head(data_new)
## Cluster gene variable value
## 1 cluster6 Unigene0604 Stage1_R1 0.8097531
## 2 cluster6 Unigene0262 Stage1_R1 0.8453759
## 3 cluster6 Unigene0069 Stage1_R1 0.8279061
## 4 cluster6 Unigene0219 Stage1_R1 0.8536163
## 5 cluster6 Unigene0116 Stage1_R1 0.8282198
## 6 cluster6 Unigene0297 Stage1_R1 0.8008572
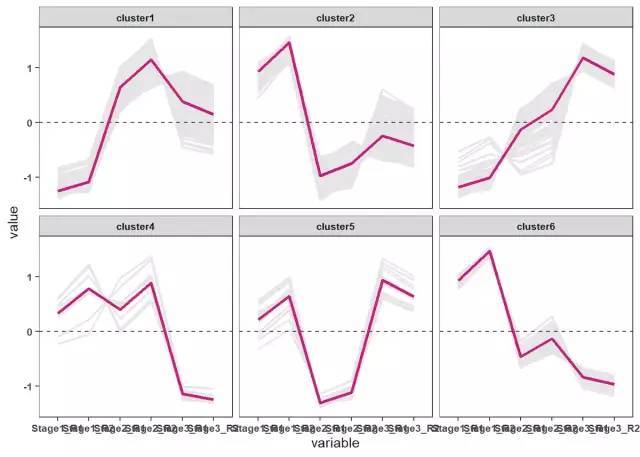
# 绘制基因表达趋势折线图
ggplot(data_new ,aes(variable ,value,group=gene ))+geom_line(color ="gray90",size =0.8)+
geom_hline(yintercept =0,linetype =2)+
stat_summary(aes(group=1),fun .y =mean ,geom ="line",size =1.2,color ="#c51b7d")+
facet_wrap(Cluster ~.)+
theme_bw()+
theme(panel .grid .major =element_blank(),panel .grid .minor =element_blank(),
axis .text =element_text(size =8,face ="bold"),
strip .text =element_text(size =8,face ="bold"))
sessionInfo
## R version 3.6.0 (2019-04-26)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 17763)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=Chinese (Simplified)_China.936
## [2] LC_CTYPE=Chinese (Simplified)_China.936
## [3] LC_MONETARY=Chinese (Simplified)_China.936
## [4] LC_NUMERIC=C
## [5] LC_TIME=Chinese (Simplified)_China.936
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] reshape2_1.4.3 pheatmap_1.0.12 ggplot2_3.2.0
##
## loaded via a namespace (and not attached):
## [1] Rcpp_1.0.1 knitr_1.23 magrittr_1.5
## [4] tidyselect_0.2.5 munsell_0.5.0 colorspace_1.4-1
## [7] R6_2.4.0 rlang_0.4.0 plyr_1.8.4
## [10] stringr_1.4.0 dplyr_0.8.3 tools_3.6.0
## [13] grid_3.6.0 gtable_0.3.0 xfun_0.8
## [16] withr_2.1.2 htmltools_0.3.6 yaml_2.2.0
## [19] lazyeval_0.2.2 digest_0.6.20 assertthat_0.2.1
## [22] tibble_2.1.3 crayon_1.3.4 RColorBrewer_1.1-2
## [25] purrr_0.3.2 glue_1.3.1 evaluate_0.14
## [28] rmarkdown_1.13 labeling_0.3 stringi_1.4.3
## [31] compiler_3.6.0 pillar_1.4.2 scales_1.0.0
## [34] pkgconfig_2.0.2